Saccharum Regulatory Networks Database
This is the database for further analysis and visualization of the data generated from "Isoform-level and haplotype-resolved full-length transcriptome revealed the lignin metabolic process in complex polyploid sugarcane cultivars". we sampled multiple tissues from the sugarcane cultivar GT42 during its whole growth period and performed full-length transcriptome sequencing (Iso-seq) and RNA sequencing (RNA-seq) to generate a multi-omics data set of sugarcane gene expression. By integrating these transcriptome datasets, we constructed an Isoform-level and haplotype-resolved full-length transcriptome of sugarcane cultivars, identifying 305,500 unique isoforms from 166,593 haplotype-resolved genes. Systematic homology inference allowed us to assign isoforms and genes to the two recognized ancestral species of sugarcane cultivars, Saccharum officinarum and Saccharum spontaneum, with approximately 28.25% of the genes derived from Saccharum spontaneum. Gene expression atlas and expression regulatory networks were used to elucidate the lignin metabolic process in sugarcane cultivars.
if you used the data from the database, please cite the paper kindly! (https://doi.org/123456/123456)


Statistics
Full-length Transcriptome
- Number of Genome: 1
- Number of Species: 4
- Number of Mosaic: 28163
- Number of Haplotype: 166593
- Number of Transcript: 305500
Variation
- SNP Type: 11
- Number of SNPs: 1310895
- ISO-Seq Evidence: 487234
- RNA-Seq Evidence: 3682156
- ISO-Seq && RNA-Seq Evidence: 442737
Gene Expression Profile
- Number of Resolutions: 4
- Development Stages: 5
- Number of Tissues: 23
- Number of Samples: 69
TF Regulatory Network
- Number of Modules: 61
- Number of Nodes: 85137
- Number of Edges: 4906837
Resources
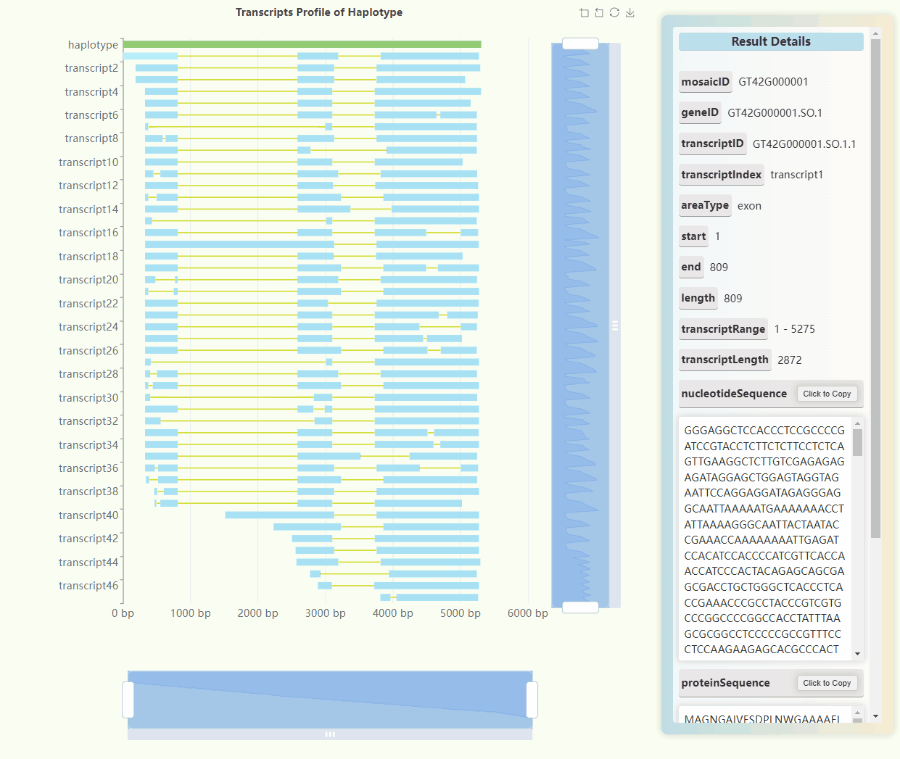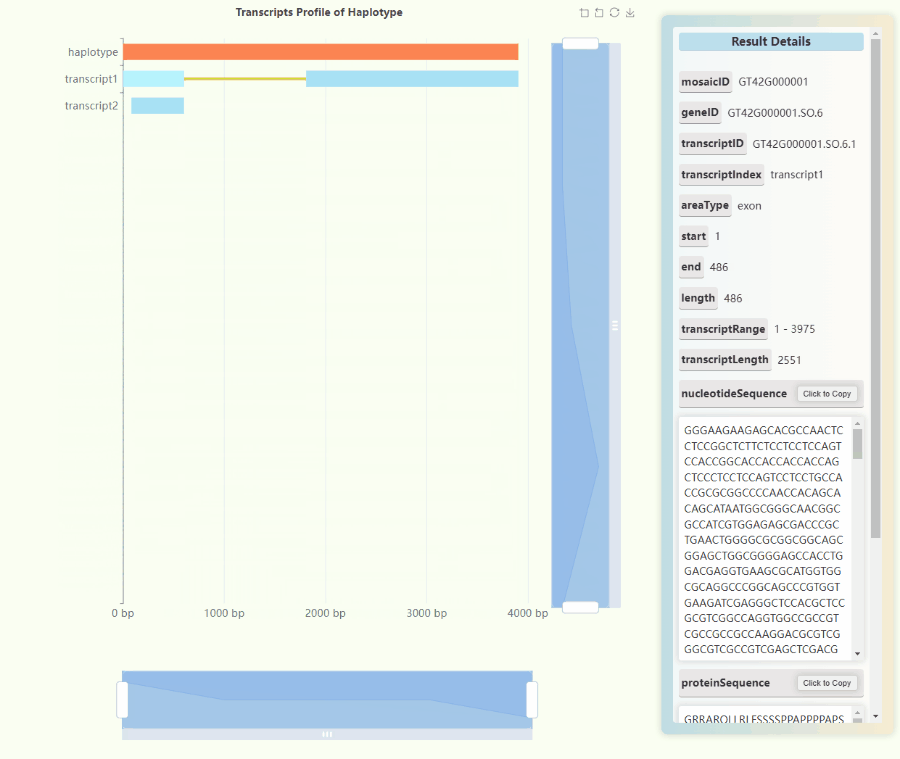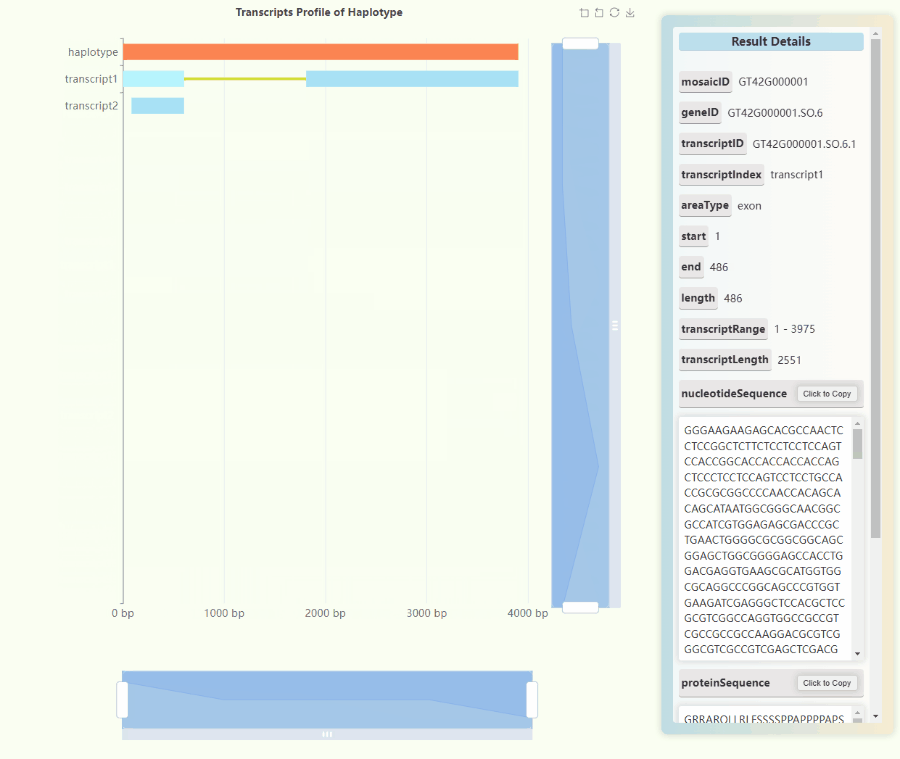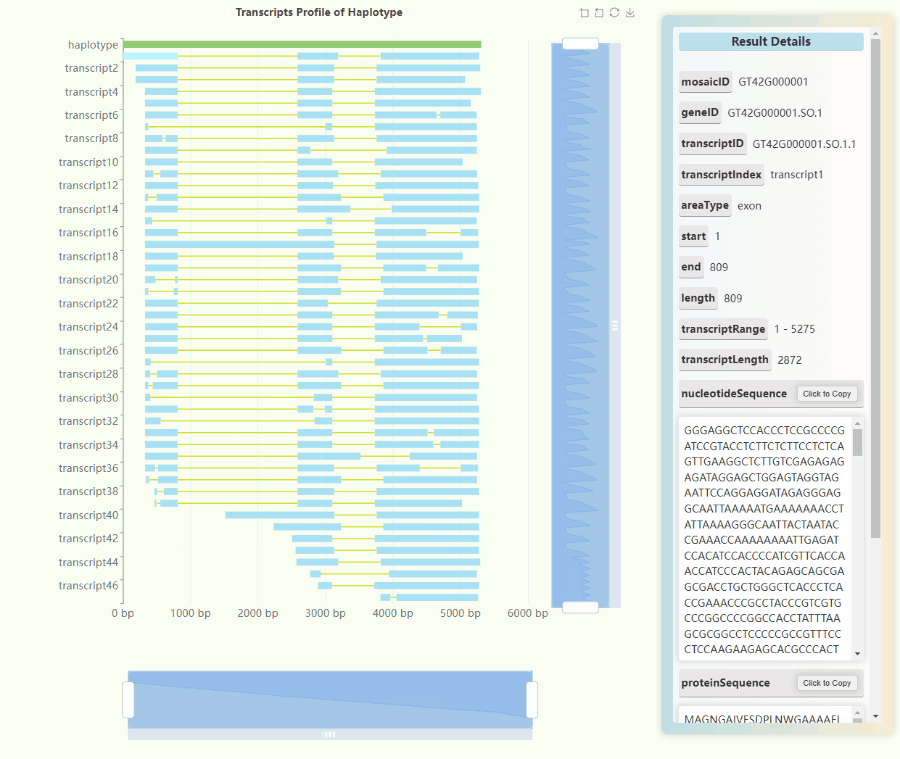
Full-length Transcriptome
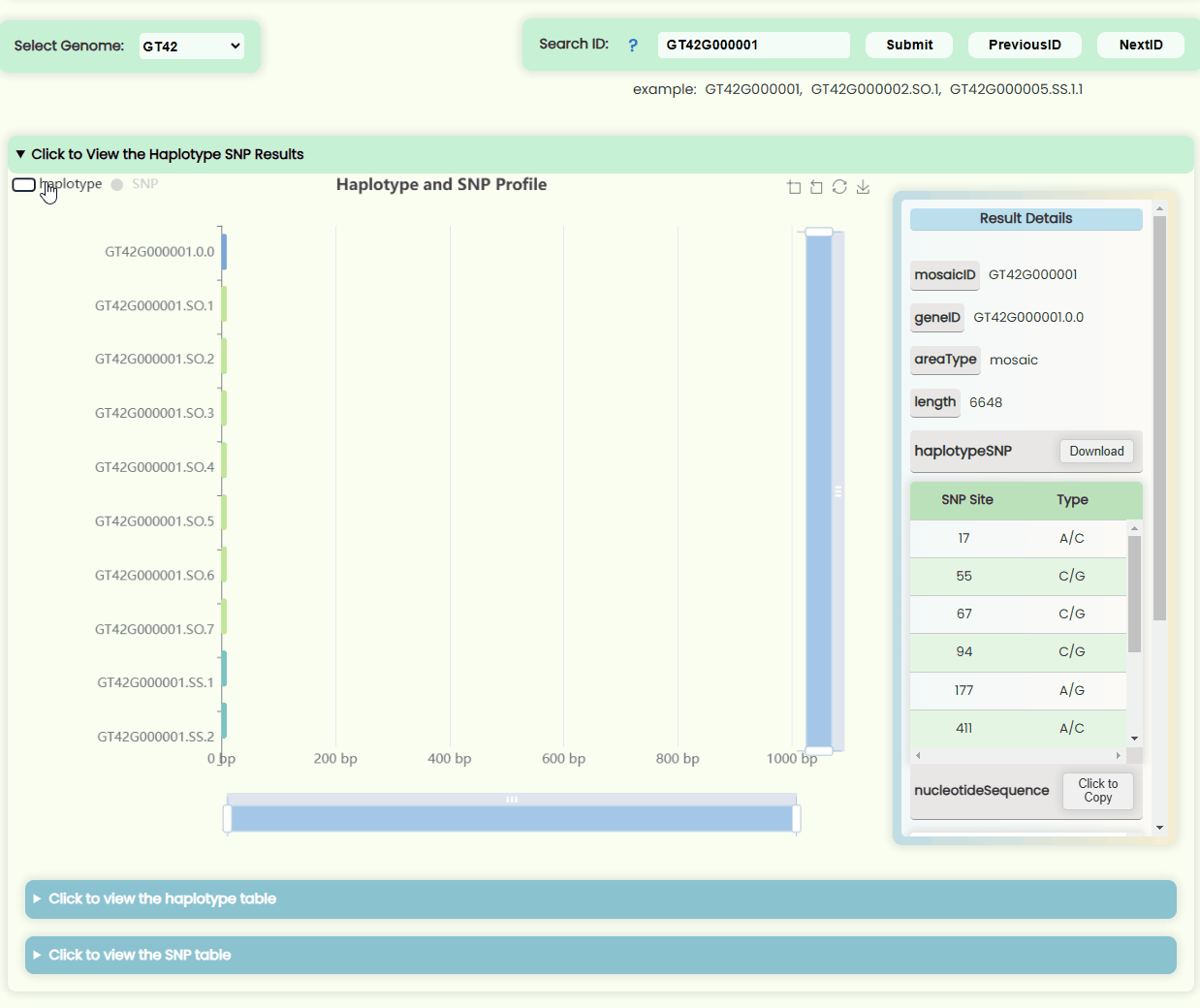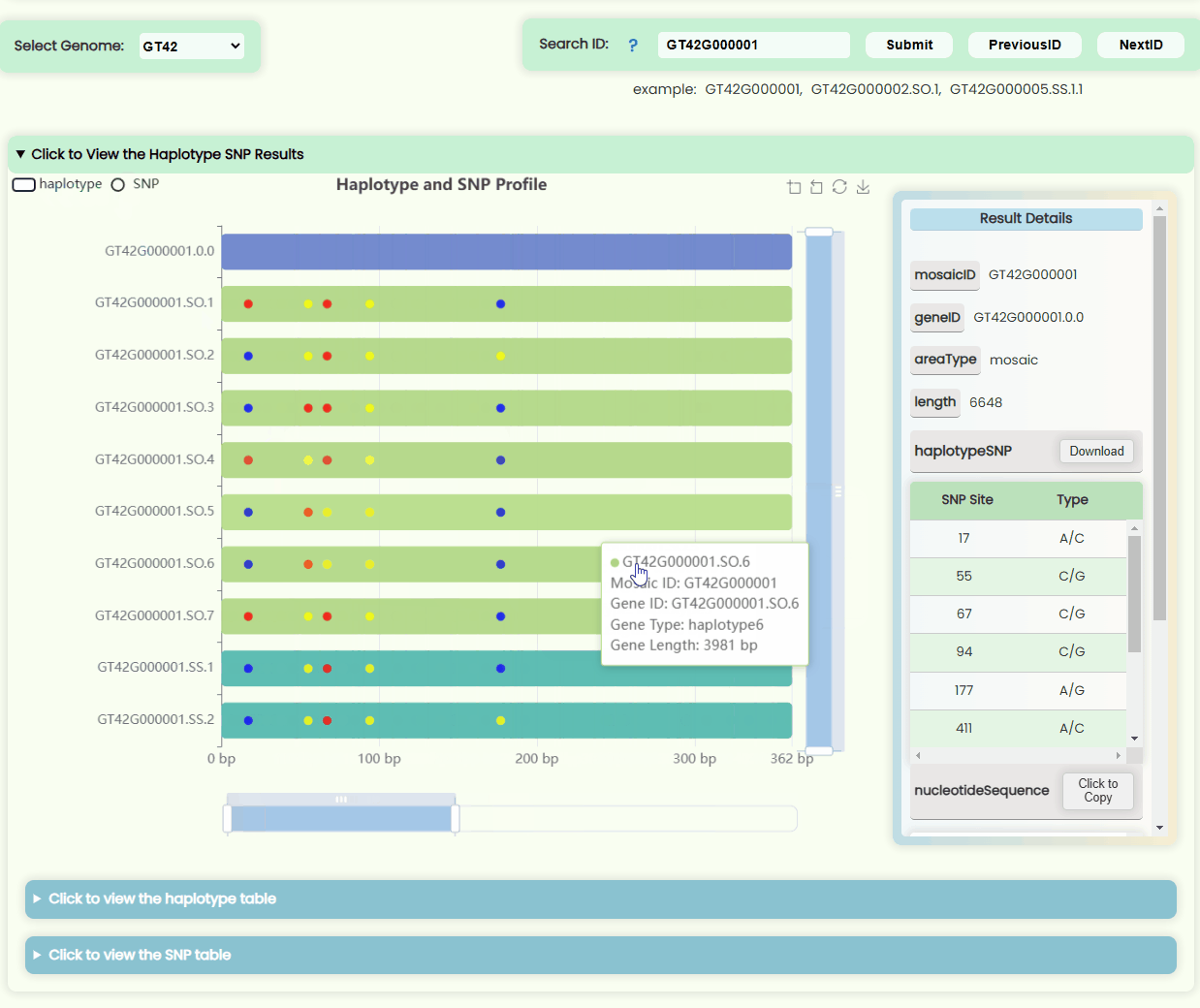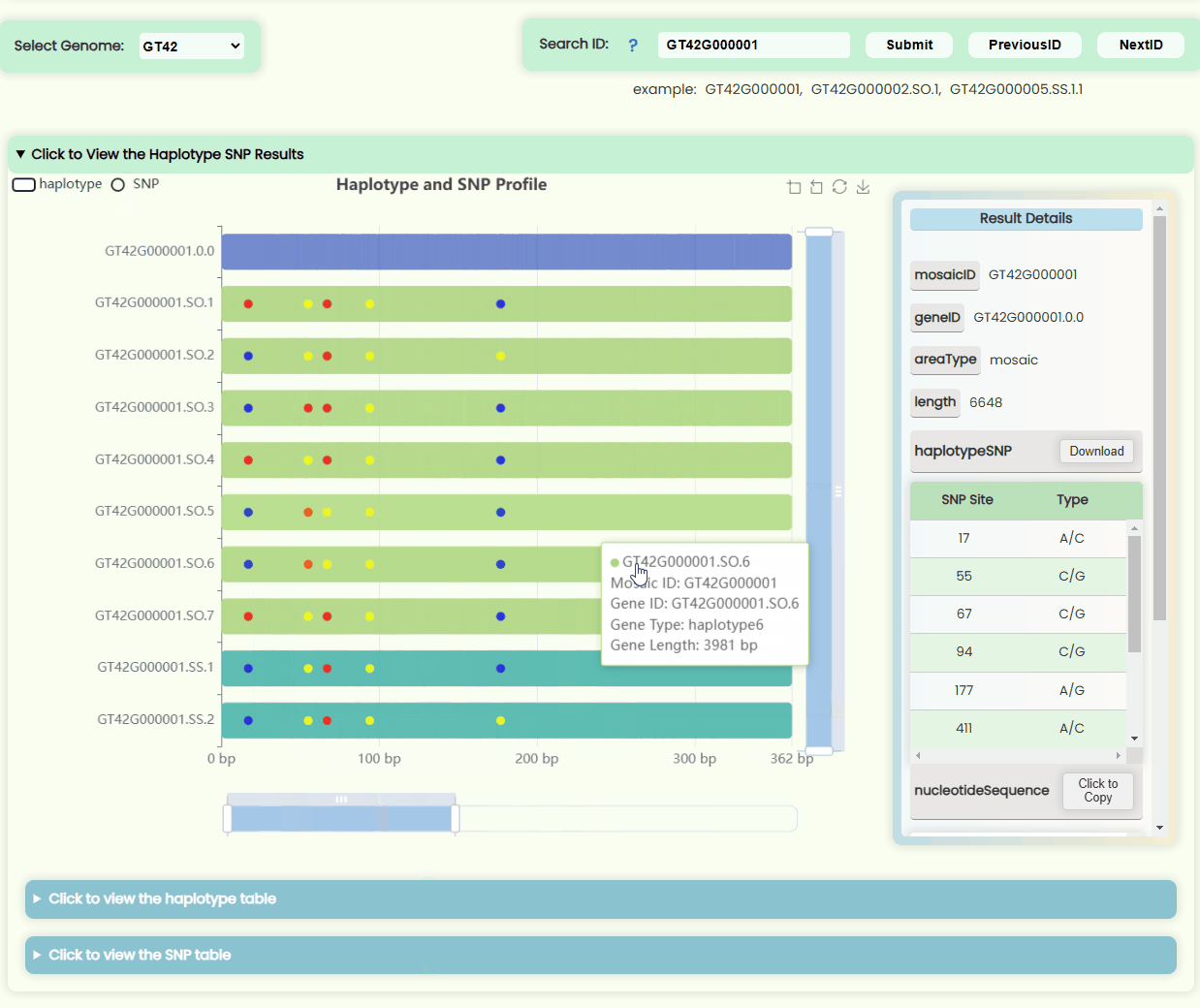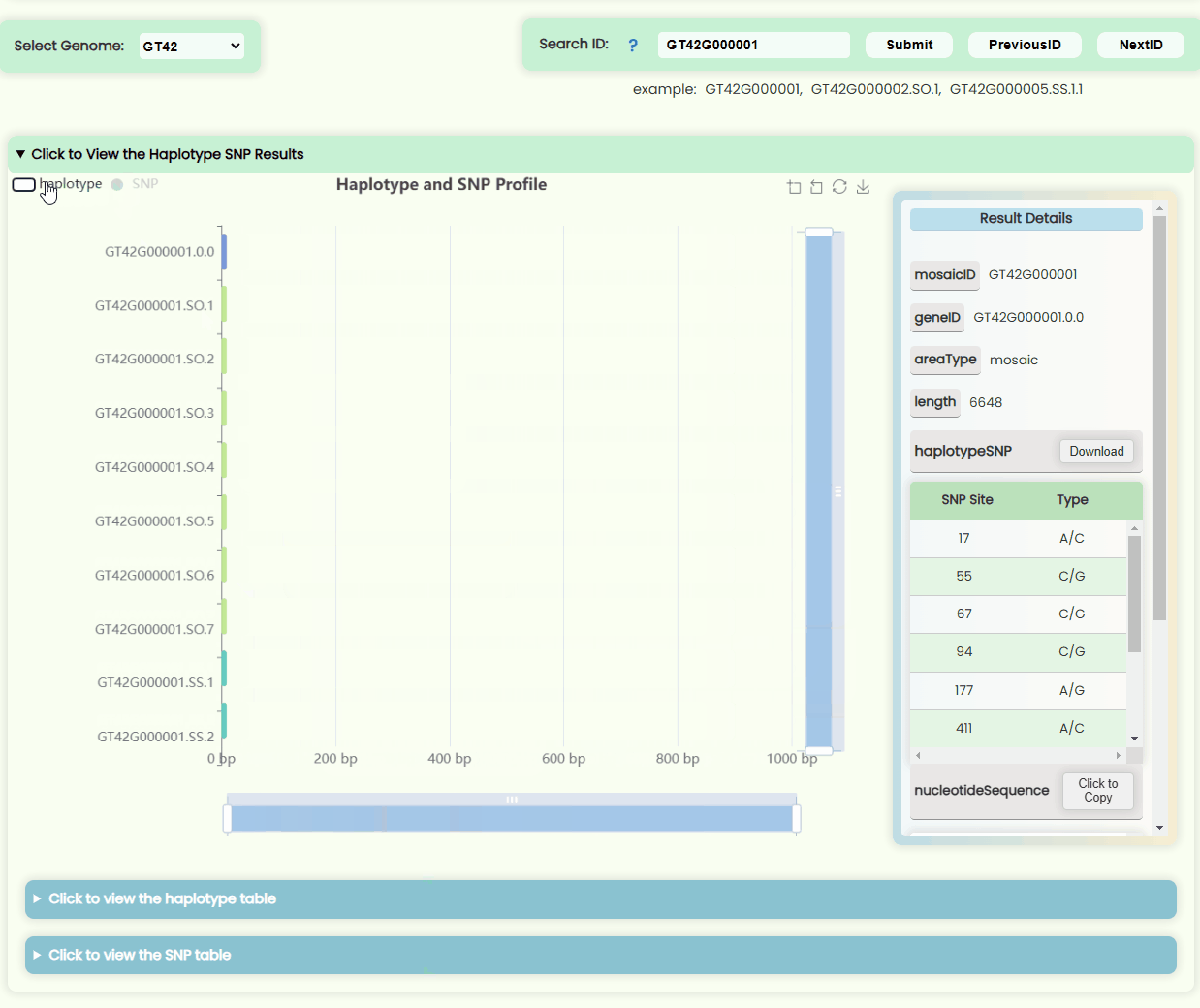
Haplotype && SNP
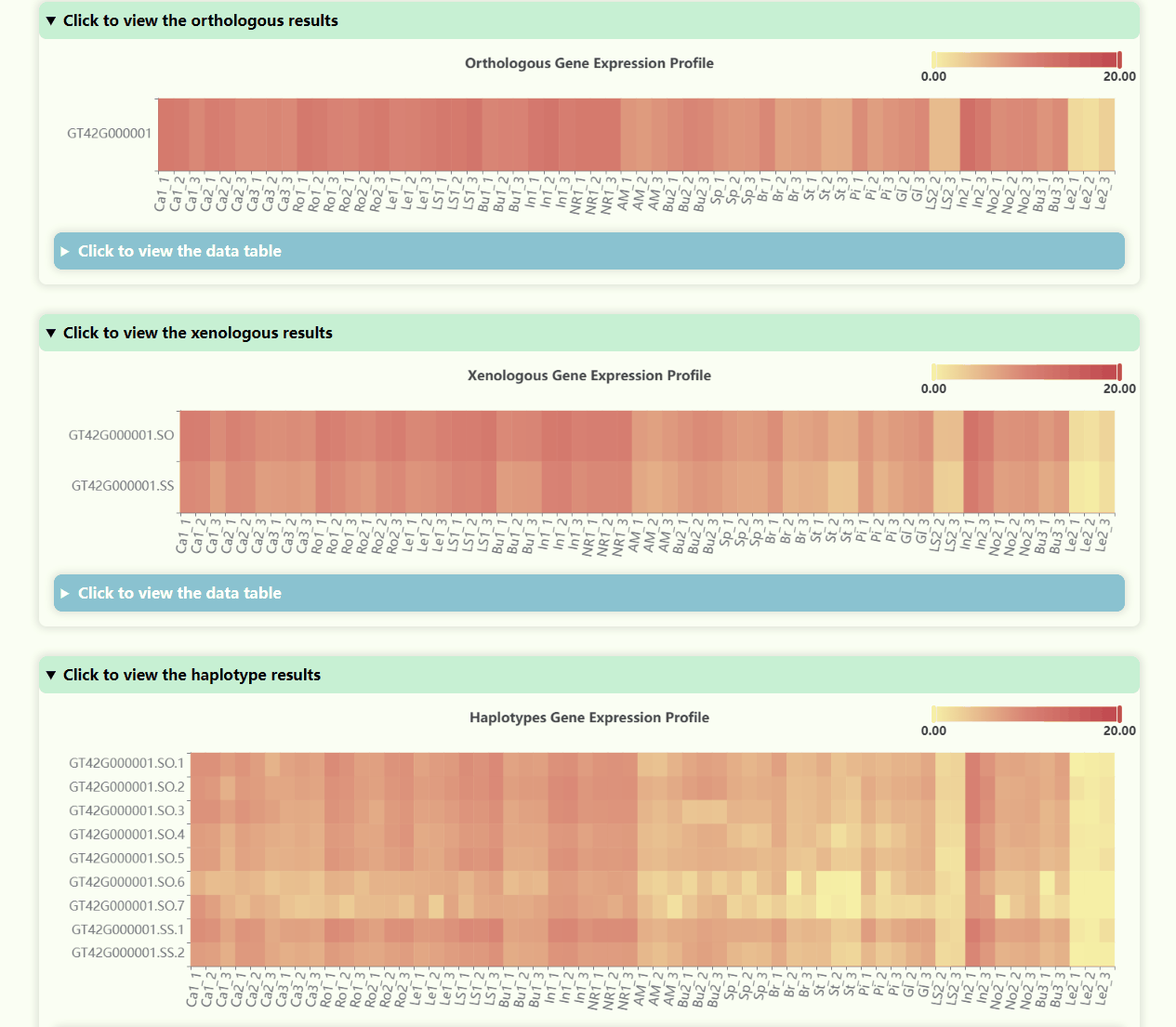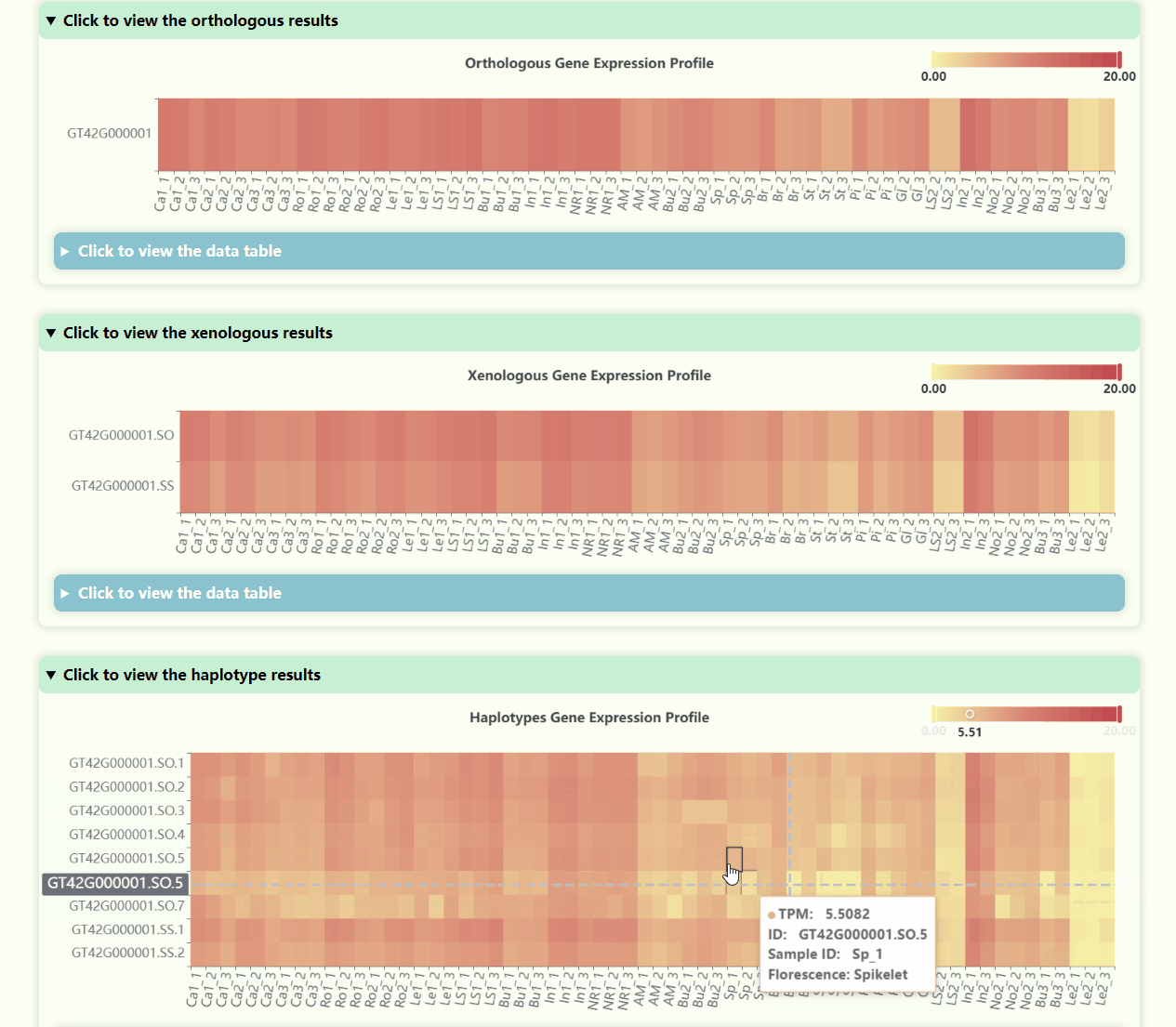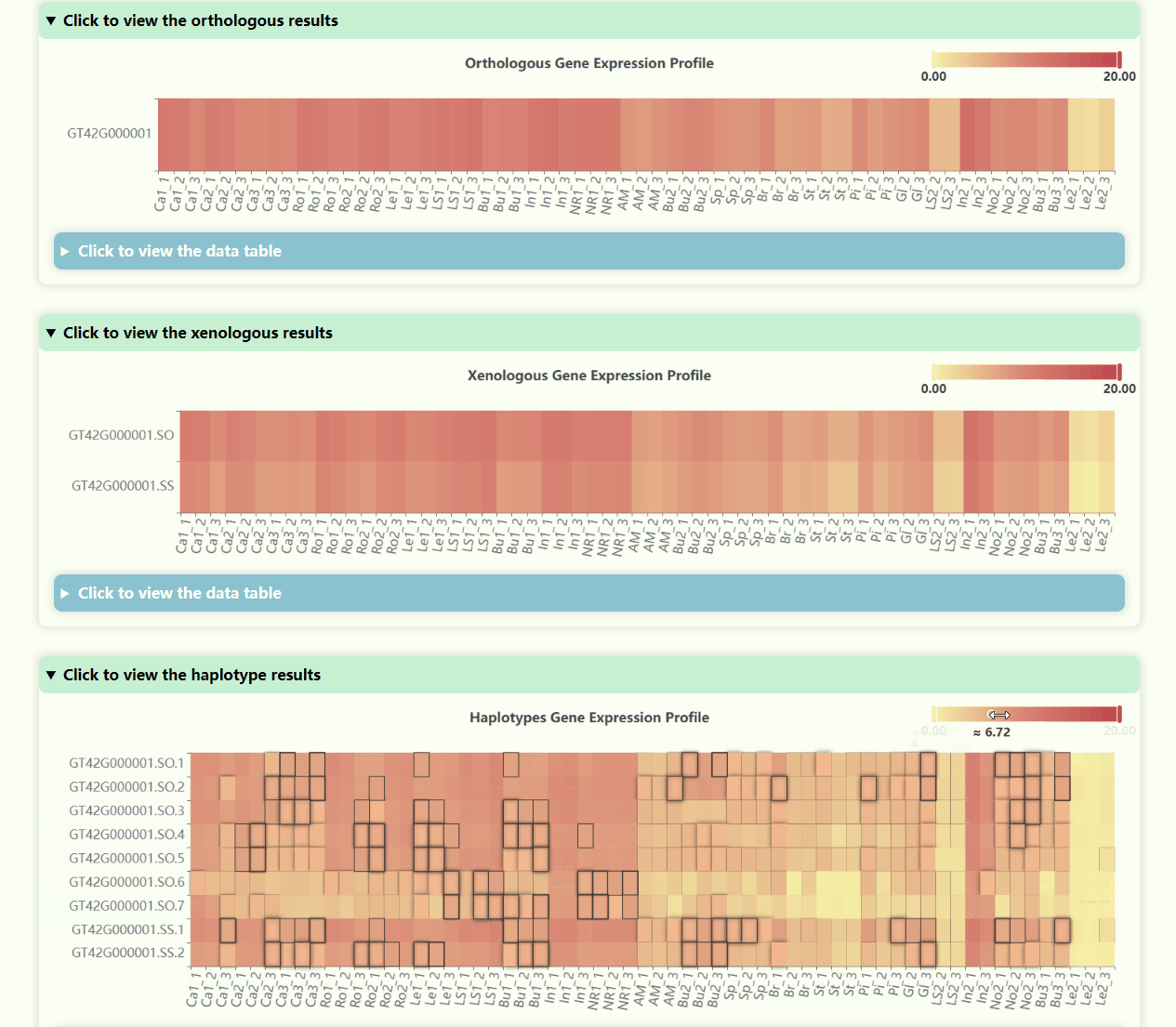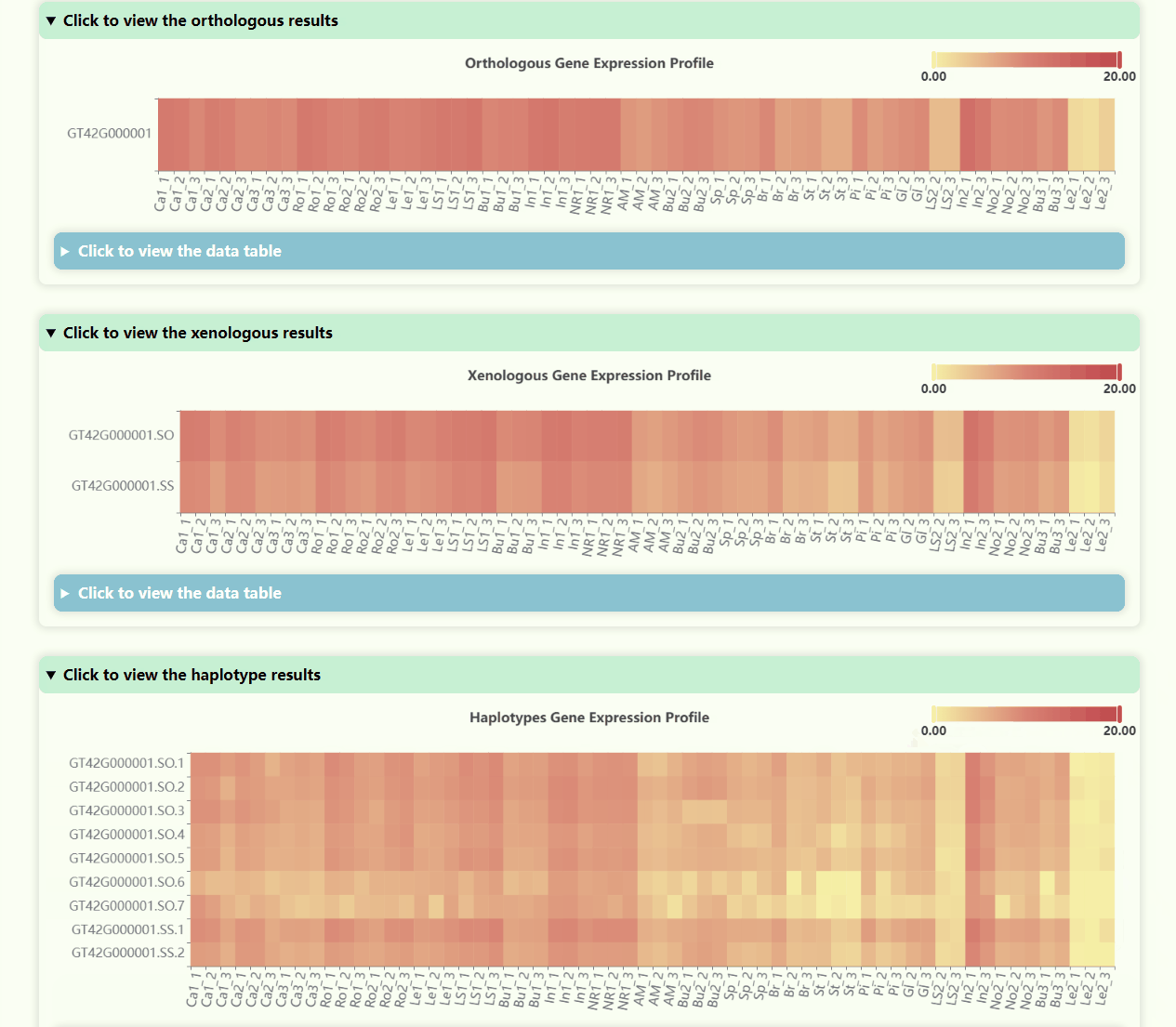
Gene Expression Profile
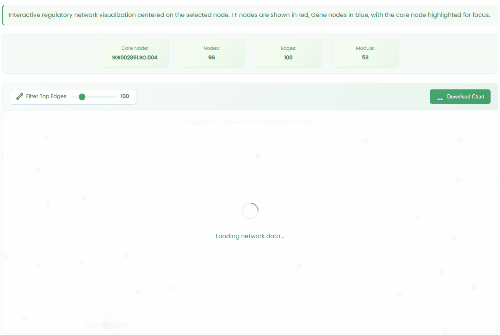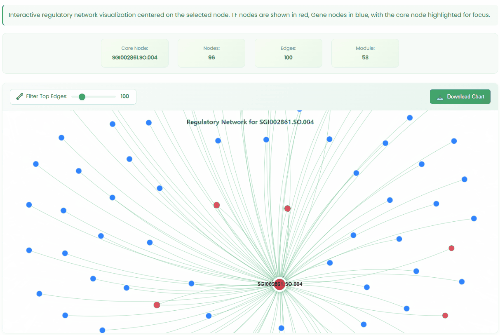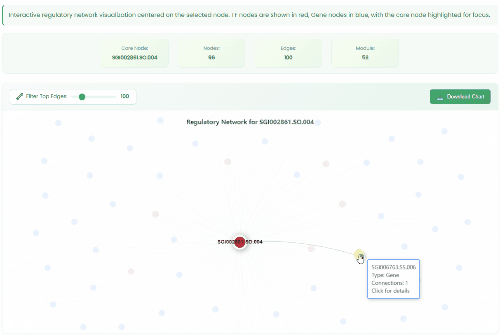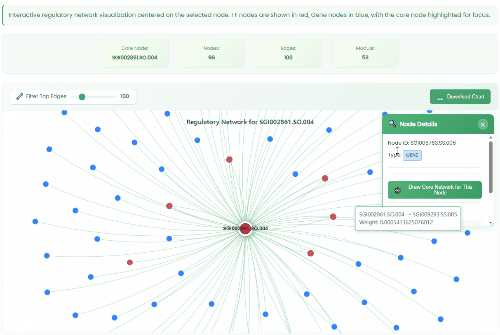
TF Regulatory Network
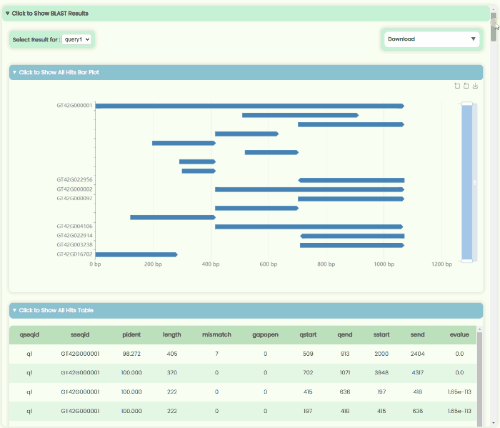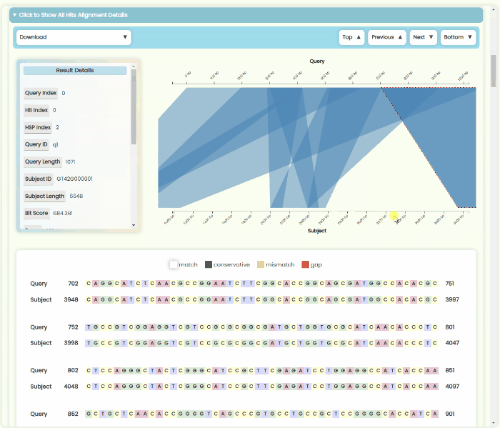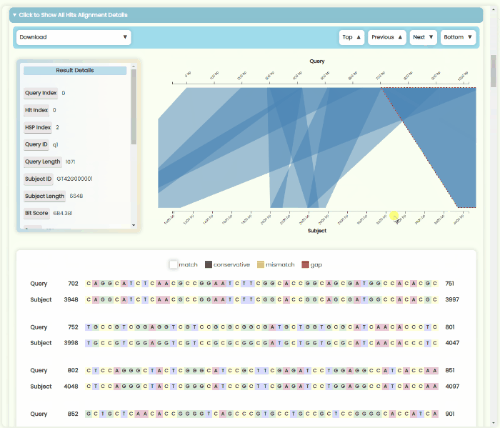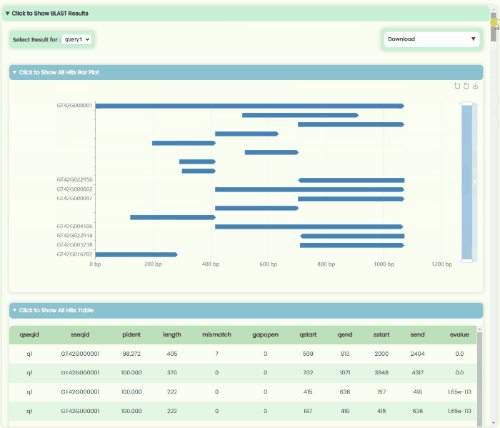
BLAST
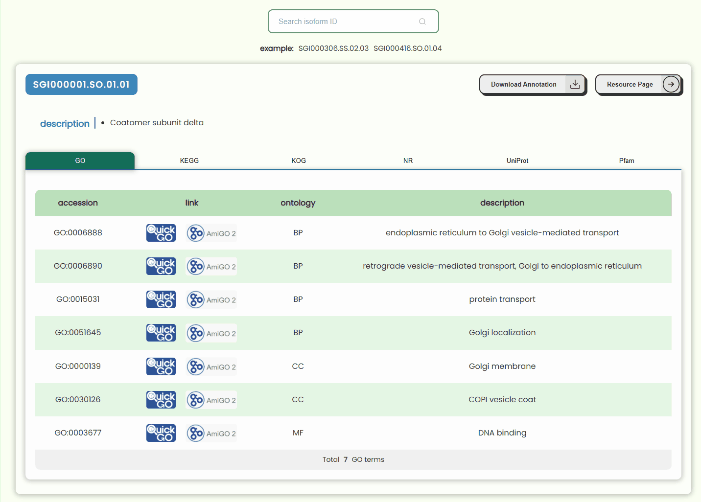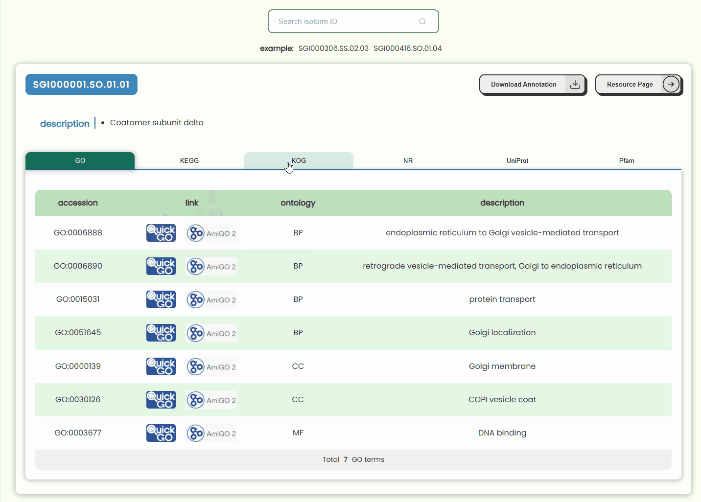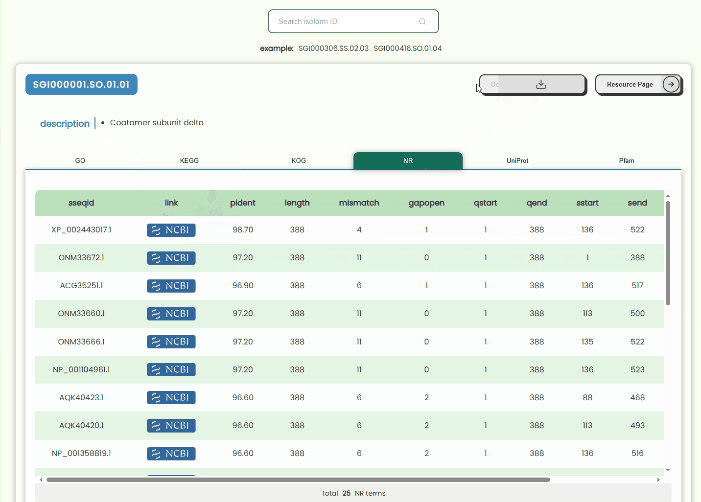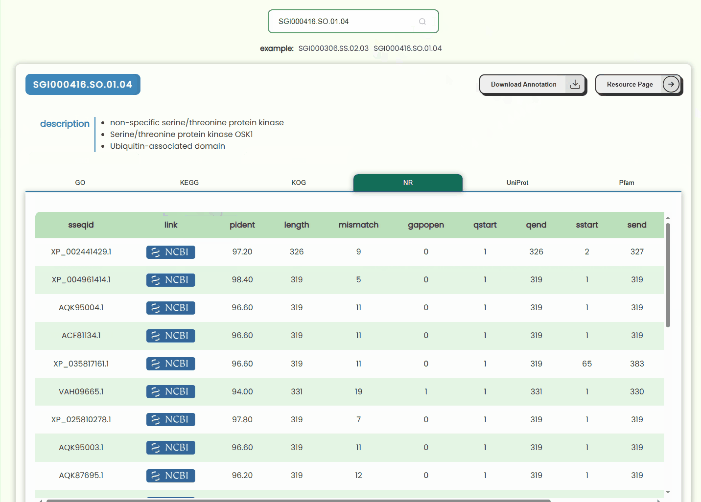
Annotation
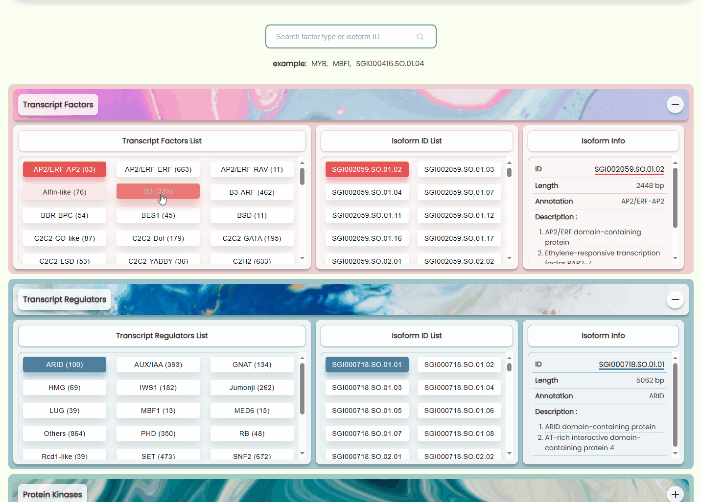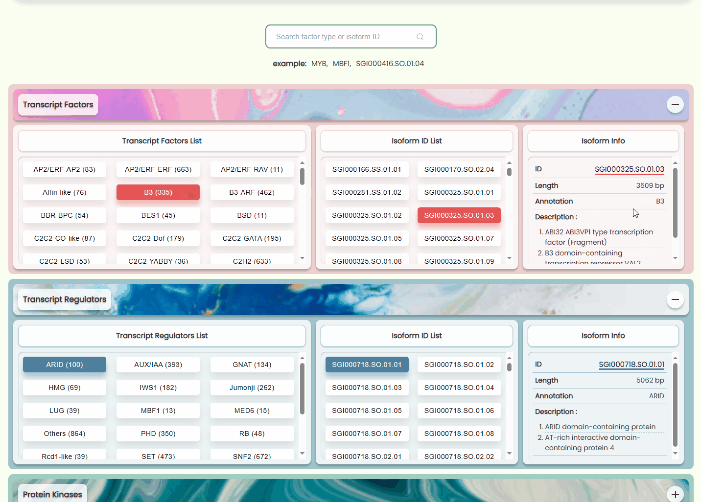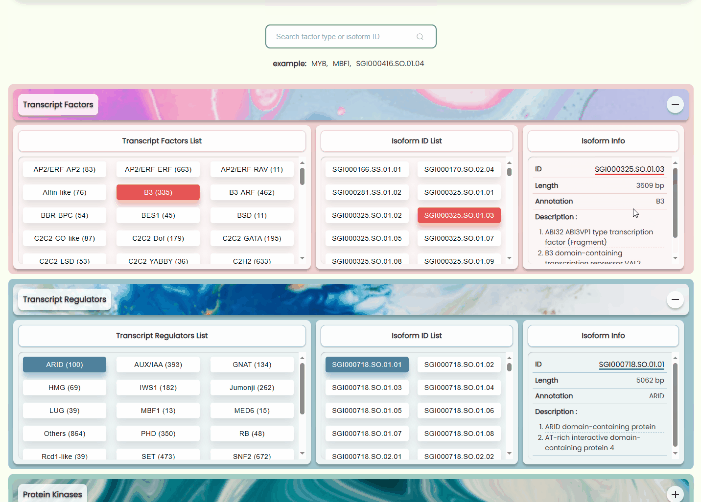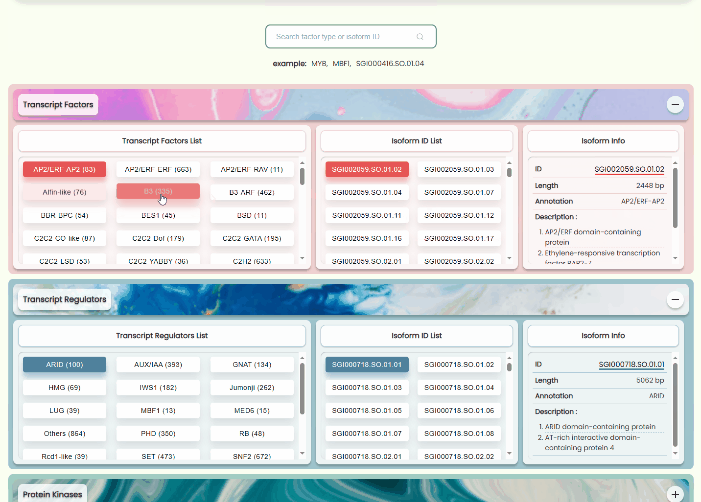
Transcript Factors
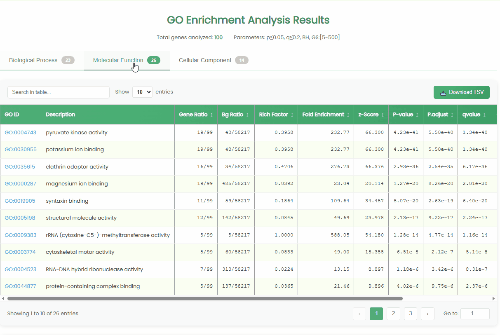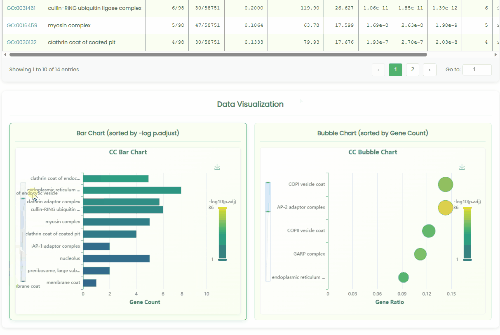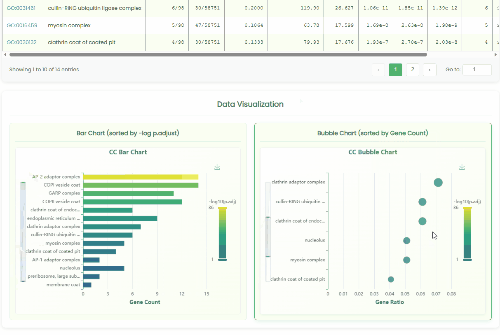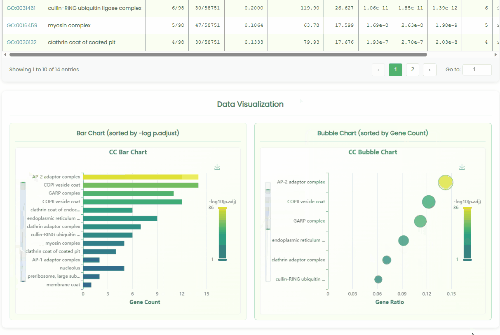
GO Enrichment
What's New
- [2025/07/23]
- [2025/01/14]
- [2024/09/21]
- [2024/06/05]
- [2024/05/12]
- [2024/02/01]
- New release of GT42_web project (v3.0.0: Infinite City) was successfully published.
- New release of GT42_web project (v2.1.0: Si Mie) was successfully published.
- Second release of GT42_web project (v2.0.0: Yu Zhe) was successfully published.
- Starting version v2.0.0 refactor.
- First release of GT42_web project (v1.0.0: Huai Yu) was successfully published.
- Starting frame design of GT42_web.


